# scRecover
*Zhun Miao, Xuegong Zhang*
*2019-04-28*

# 1. Introduction
**`scRecover`** is an R package for **imputation of single-cell RNA-seq (scRNA-seq) data**. It will detect and impute dropout values in a scRNA-seq raw read counts matrix while **keeping the real zeros unchanged**.
Since there are both dropout zeros and real zeros in scRNA-seq data, imputation methods should not impute all the zeros to non-zero values. To distinguish dropout and real zeros, **`scRecover`** employs the Zero-Inflated Negative Binomial (ZINB) model for dropout probability estimation of each gene and accumulation curves for prediction of dropout number in each cell. By combination with scImpute, SAVER and MAGIC, it not only detects dropout and real zeros **at higher accuracy**, but also **improve the downstream clustering and visualization results**.
# 2. Citation
If you use **`scRecover`** in published research, please cite:
>
# 3. Installation
To install **`scRecover`** from [**Bioconductor**](http://bioconductor.org/packages/scRecover/):
```{r Installation from Bioconductor, eval = FALSE}
if(!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("scRecover")
```
To install the *developmental version* from [**Bioconductor**](https://bioconductor.org/packages/devel/bioc/html/scRecover.html):
```{r Developmental version from Bioconductor, eval = FALSE}
if(!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("scRecover", version = "devel")
```
Or install the *developmental version* from [**GitHub**](https://github.com/miaozhun/scRecover/):
```{r Installation from GitHub, eval = FALSE}
if(!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("miaozhun/scRecover")
```
To load **`scRecover`** and other required packages for the vignettes in R:
```{r Load scRecover, eval = TRUE}
library(scRecover)
library(BiocParallel)
suppressMessages(library(SingleCellExperiment))
```
# 4. Input
**`scRecover`** takes two inputs: `counts` and one of `Kcluster` or `labels`.
The input `counts` is a scRNA-seq **read counts matrix** or a **`SingleCellExperiment`** object which contains the read counts matrix. The rows of the matrix are genes and columns are cells.
`Kcluster` is an integer specifying the number of cell subpopulations. This parameter can be determined based on prior knowledge or clustering of raw data. `Kcluster` is used to determine the candidate neighbors of each cell and need not to be very accurate.
`labels` is a character/integer vector specifying the cell type of each column in the raw count matrix. Only needed when `Kcluster = NULL`. Each cell type should have at least two cells for imputation.
# 5. Test data
Users can load the test data in **`scRecover`** by
```{r Load scRecoverTest}
data(scRecoverTest)
```
The test data `counts` in `scRecoverTest` is a scRNA-seq read counts matrix which has 200 genes (rows) and 150 cells (columns).
```{r counts}
dim(counts)
counts[1:6, 1:6]
```
The object `labels` in `scRecoverTest` is a vector of integer specifying the cell types in the read counts matrix, corresponding to the columns of `counts`.
```{r labels}
length(labels)
table(labels)
```
The object `oneCell` in `scRecoverTest` is a vector of a cell's raw read counts for each gene.
```{r oneCell}
head(oneCell)
length(oneCell)
```
# 6. Usage
## 6.1 Imputation using scRecover
### 6.1.1 With read counts matrix input
Here is an example to run **`scRecover`** with read counts matrix input:
```{r demo1, eval = TRUE}
# Load test data for scRecover
data(scRecoverTest)
# Run scRecover with Kcluster specified
scRecover(counts = counts, Kcluster = 2, outputDir = "./outDir_scRecover/", verbose = FALSE)
# Or run scRecover with labels specified
# scRecover(counts = counts, labels = labels, outputDir = "./outDir_scRecover/")
```
### 6.1.2 With SingleCellExperiment input
The [`SingleCellExperiment`](http://bioconductor.org/packages/SingleCellExperiment/) class is a widely used S4 class for storing single-cell genomics data. **`scRecover`** also could take the `SingleCellExperiment` data representation as input.
Here is an example to run **`scRecover`** with `SingleCellExperiment` input:
```{r demo2, eval = TRUE}
# Load test data for scRecover
data(scRecoverTest)
# Convert the test data in scRecover to SingleCellExperiment data representation
sce <- SingleCellExperiment(assays = list(counts = as.matrix(counts)))
# Run scRecover with SingleCellExperiment input sce (Kcluster specified)
scRecover(counts = sce, Kcluster = 2, outputDir = "./outDir_scRecover/", verbose = FALSE)
# Or run scRecover with SingleCellExperiment input sce (labels specified)
# scRecover(counts = sce, labels = labels, outputDir = "./outDir_scRecover/")
```
## 6.2 Estimate dropout gene number in a cell
Function `estDropoutNum` in the package could estimate the dropout gene number or all expressed gene number (namely observed gene number plus dropout gene number) in a cell:
```{r demo3, eval = TRUE}
# Load test data
data(scRecoverTest)
# Downsample 10% read counts in oneCell
set.seed(999)
oneCell.down <- countsSampling(counts = oneCell, fraction = 0.1)
# Count the groundtruth dropout gene number in the downsampled cell
sum(oneCell.down == 0 & oneCell != 0)
# Estimate the dropout gene number in the downsampled cell by estDropoutNum
estDropoutNum(sample = oneCell.down, depth = 10, return = "dropoutNum")
```
Blow shows the expressed gene number predicted by `estDropoutNum` with 10% downsampled reads and the groundtruth expressed gene number derived by downsampling when the reads depth varying from 0% to 100% of the total reads.

# 7. Output
Imputed expression matrices of **`scRecover`** will be saved in the output directory specified by \code{outputDir} or a folder named with prefix 'outDir_scRecover_' under the current working directory when \code{outputDir} is unspecified.
# 8. Parallelization
**`scRecover`** integrates parallel computing function with [`BiocParallel`](http://bioconductor.org/packages/BiocParallel/) package. Users could just set `parallel = TRUE` (default) in function `scRecover` to enable parallelization and leave the `BPPARAM` parameter alone.
```{r demo4, eval = FALSE}
# Run scRecover with Kcluster specified
scRecover(counts = counts, Kcluster = 2, parallel = TRUE)
# Run scRecover with labels specified
scRecover(counts = counts, labels = labels, parallel = TRUE)
```
Advanced users could use a `BiocParallelParam` object from package `BiocParallel` to fill in the `BPPARAM` parameter to specify the parallel back-end to be used and its configuration parameters.
## 8.1 For Unix and Mac users
The best choice for Unix and Mac users is to use `MulticoreParam` to configure a multicore parallel back-end:
```{r demo5, eval = FALSE}
# Set the parameters and register the back-end to be used
param <- MulticoreParam(workers = 18, progressbar = TRUE)
register(param)
# Run scRecover with 18 cores (Kcluster specified)
scRecover(counts = counts, Kcluster = 2, parallel = TRUE, BPPARAM = param)
# Run scRecover with 18 cores (labels specified)
scRecover(counts = counts, labels = labels, parallel = TRUE, BPPARAM = param)
```
## 8.2 For Windows users
For Windows users, use `SnowParam` to configure a Snow back-end is a good choice:
```{r demo6, eval = FALSE}
# Set the parameters and register the back-end to be used
param <- SnowParam(workers = 8, type = "SOCK", progressbar = TRUE)
register(param)
# Run scRecover with 8 cores (Kcluster specified)
scRecover(counts = counts, Kcluster = 2, parallel = TRUE, BPPARAM = param)
# Run scRecover with 8 cores (labels specified)
scRecover(counts = counts, labels = labels, parallel = TRUE, BPPARAM = param)
```
See the [*Reference Manual*](https://bioconductor.org/packages/release/bioc/manuals/BiocParallel/man/BiocParallel.pdf) of [`BiocParallel`](http://bioconductor.org/packages/BiocParallel/) package for more details of the `BiocParallelParam` class.
# 9. Evaluation of scRecover
## 9.1 On downsampling data
We evaluated SAVER, scImpute, MAGIC and their combined with scRecover version SAVER+scRecover, scImpute+scRecover, MAGIC+scRecover on a downsampling scRNA-seq dataset generated by random sampling of reads from a SMART-seq2 scRNA-seq dataset (Petropoulos S, et al. Cell, 2016, https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-3929/).
### 9.1.1 Accuracy of dropout prediction
We found after combined with scRecover, scImpute+scRecover, SAVER+scRecover and MAGIC+scRecover will have higher accuracy than scImpute, SAVER and MAGIC respectively.

### 9.1.2 Predicted dropout number
We found scImpute+scRecover, SAVER+scRecover and MAGIC+scRecover will have predicted dropout numbers closer to the real dropout number than without combination with scRecover.

## 9.2 On 10X data
We applied the 6 imputation methods to a 10X scRNA-seq dataset (https://support.10xgenomics.com/single-cell-gene-expression/datasets/3.0.0/heart_1k_v3).
Then we measured the downstream clustering and visualization results by comparing to the cell labels originated from the dataset and deriving their Adjusted Rand Index (ARI) and Jaccard indexes (the larger, the better).
We found a significant improvement of SAVER, scImpute and MAGIC after combined with scRecover.

Gene number before and after imputation:
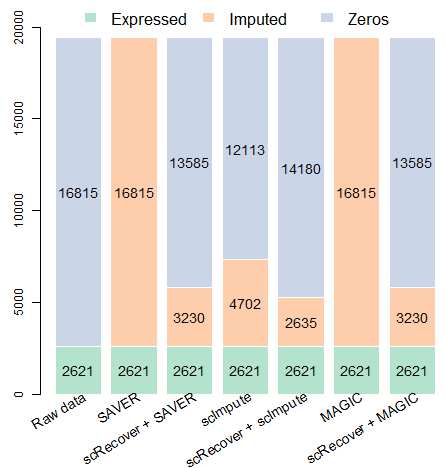
## 9.3 On SMART-seq data
Next, we applied the 6 imputation methods to a SMART-seq scRNA-seq dataset (Chu L, et al. Genome Biology, 2016, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE75748).
Then we measured the downstream clustering and visualization results by comparing to the cell labels originated from the dataset and deriving their Adjusted Rand Index (ARI) and Jaccard indexes (the larger, the better).
We found a significant improvement of SAVER, scImpute and MAGIC after combined with scRecover.

Gene number before and after imputation:

# 10. Help
Use `browseVignettes("scRecover")` to see the vignettes of **`scRecover`** in R after installation.
Use the following code in R to get access to the help documentation for **`scRecover`**:
```{r help, eval = FALSE}
# Documentation for scRecover
?scRecover
# Documentation for estDropoutNum
?estDropoutNum
# Documentation for countsSampling
?countsSampling
# Documentation for normalization
?normalization
# Documentation for test data
?scRecoverTest
?counts
?labels
?oneCell
```
You are also welcome to contact the author by email for help.
# 11. Author
*Zhun Miao, Xuegong Zhang* <<zhangxg@tsinghua.edu.cn>>
MOE Key Laboratory of Bioinformatics; Bioinformatics Division and Center for Synthetic & Systems Biology, TNLIST; Department of Automation, Tsinghua University, Beijing 100084, China.