# scFeatures: Multi-view representations of single-cell and spatial data for disease outcome prediction
<img src="https://raw.githubusercontent.com/SydneyBioX/scFeatures/devel/inst/sticker.png" align="right" width="200">
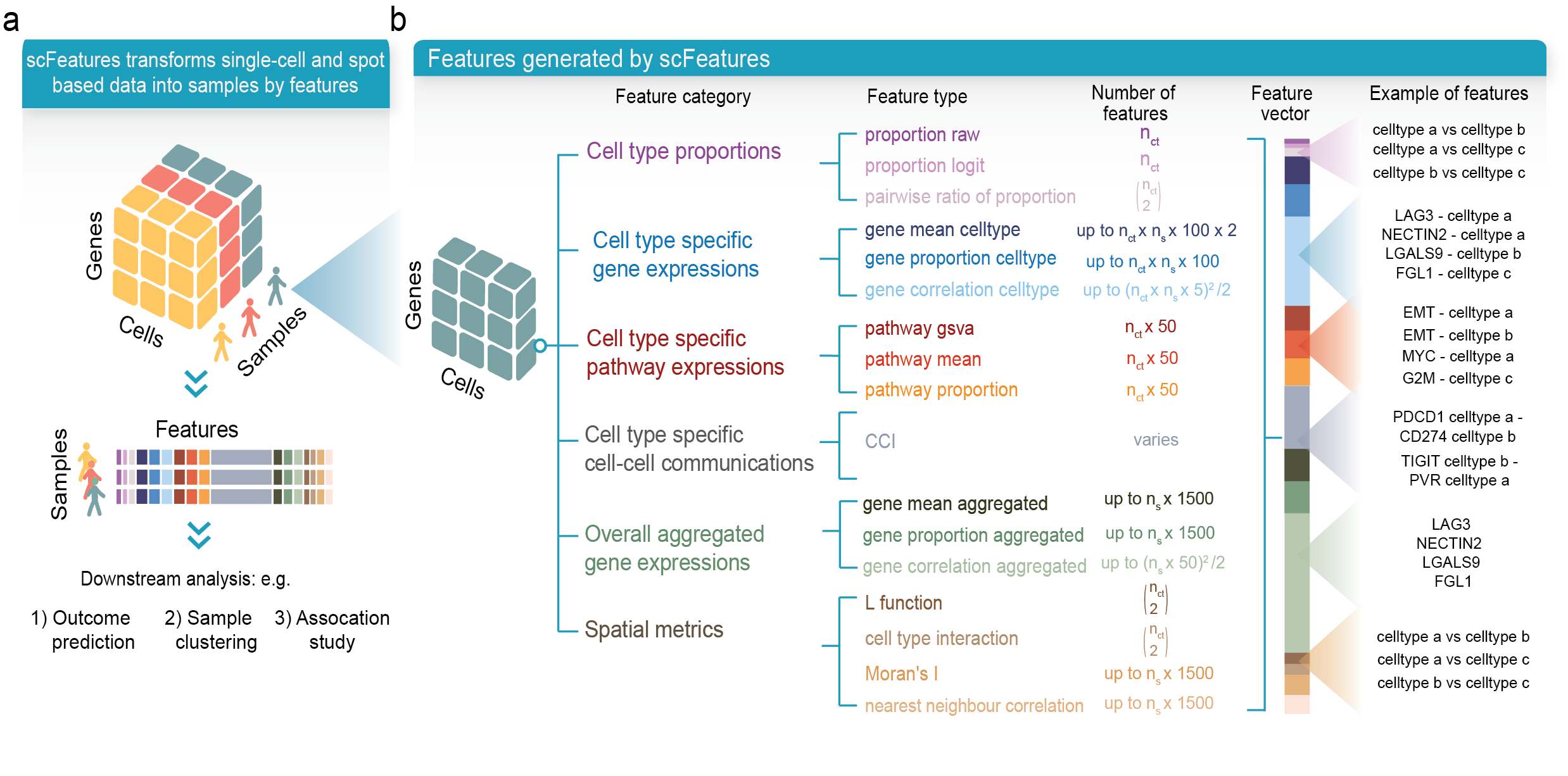
scFeatures is a tool that generates multi-view representations of single-cell and spatial data through the construction of a total of 17 feature types belonging to the following six categories.
1. cell type proportions
2. cell type specific gene expressions
3. cell type specific pathway expressions
4. cell type specific cell-cell interaction (CCI) scores
5. overall aggregated gene expressions
6. spatial metrics
## Installation
The latest scFeatures can be installed using devtools:
```
library(devtools)
devtools::install_github("SydneyBioX/scFeatures")
```
## Vignettes
Please see https://sydneybiox.github.io/scFeatures/articles/scFeatures_overview.html.
## Reference
Cao, Y., Lin, Y., Patrick, E., Yang, P., & Yang, J. Y. H. (2022). scFeatures: multi-view representations of single-cell and spatial data for disease outcome prediction. In O. Vitek (Ed.), Bioinformatics (Vol. 38, Issue 20, pp. 4745–4753). Oxford University Press (OUP). https://doi.org/10.1093/bioinformatics/btac590