# gDR overview
gDR is an umbrella package for the gDR programmatic R interface.
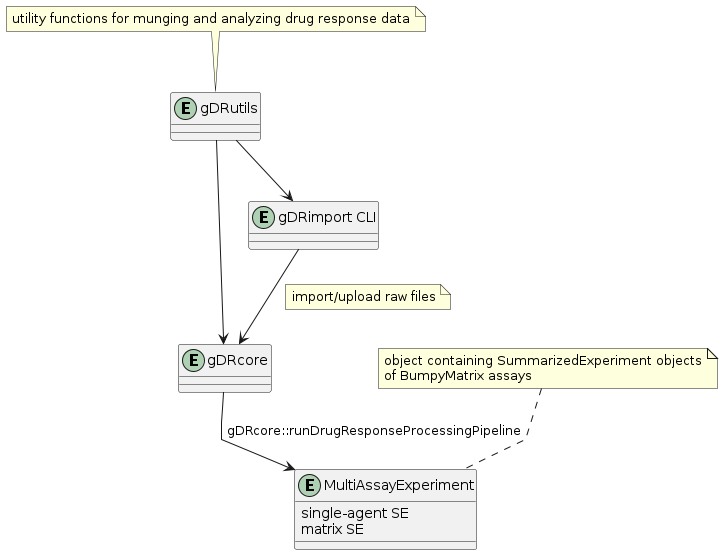
The gDR suite offers a full stack solution for processing drug response data. This enables a range of users across computational savvy, (i.e. lab scientists and computational scientists alike) to access the same, standardized data. The suite is made up of several core R packages.
# Website
A package website is available under [this link](https://gdrplatform.github.io/gDR/).
# Installation
The easiest way to use functions from gDR package is by using the Docker image. Here is one of the ways to do that.
1. Generate new personal token
Firstly, generate new personal token if you don't have one (go to https://github.com/settings/tokens/new).
2. Clone the gDR repository
Then, clone the gDR repository via https (not ssh). Please use the personal token when asked for the password.
```
git clone https://github.com/gdrplatform/gDR.git
```
3. [Optional] Save access token into .github_access_token.txt file
Write your access token as a single line in the file: `rplatform/.github_access_token.txt`.
```
cp .github_access_token.txt rplatform/
```
4. Docker build
Then build the Docker image.
```
bash extras/create_image.sh
```
5. Create 'gdr_rstudio' container with RStudio IDE available at http://localhost:8787 (default credentials are login: `rstudio` and password: `gdr`).
```
bash extras/create_container.sh
```
You've just successfully set up the environment for the gDR umbrella package. Please see [gDR vignette](vignettes/gDR.Rmd) for examples of usage.
# How to contribute
We are eager to make this tool useful for the community. We welcome any suggestions via GitHub issues, or pull requests for bugfixes/new features. When making pull requests, please first have a look at the style conventions in [gDRstyle](https://gdrplatform.github.io/gDRstyle/style_guide.html).
# Preparing your machine to run gDR images
1. How to install Docker.
Install Docker on MacOS
https://docs.docker.com/desktop/install/mac-install/
Install Docker on Windows
https://docs.docker.com/desktop/install/windows-install/
Install Docker on Linux
https://docs.docker.com/desktop/install/linux-install/
2. MacOS troubleshooting (M1/M2)
```
Could not connect to the R session on RStudio Server
Unable to connect to service (1)
```
To solve the above problem, please execute the steps below:
```
Go to Settings
```
```
Click Features in development
```
```
Select checkbox: Use Rossetta for x86/amd64 emulation on Apple Silicon
```