## PrInCE
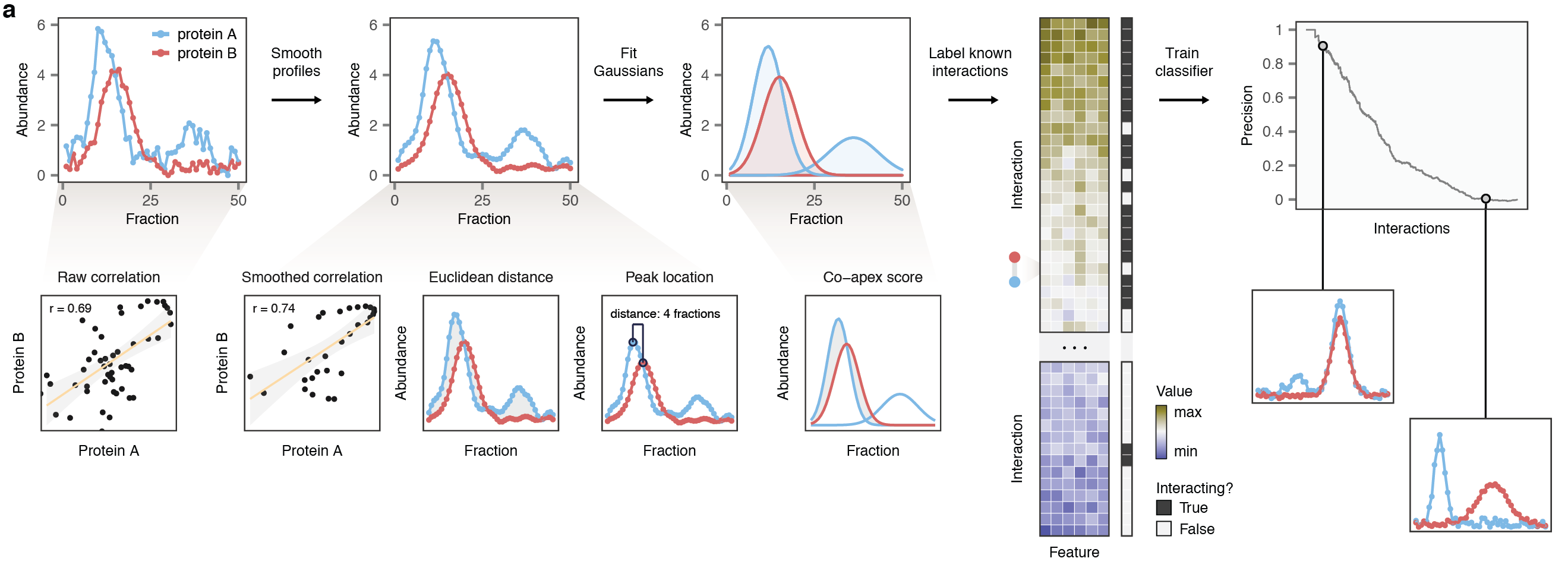
PrInCE is a computational approach to infer protein-protein interaction networks from co-elution proteomics data, also called co-migration, co-fractionation, or protein correlation profiling. This family of methods separates interacting protein complexes on the basis of their diameter or biochemical properties. Protein-protein interactions can then be inferred for pairs of proteins with similar elution profiles. PrInCE implements a machine-learning approach to identify protein-protein interactions given a set of labelled examples, using features derived exclusively from the data. This allows PrInCE to infer high-quality protein interaction networks from raw proteomics data, without bias towards known interactions or functionally associated proteins, making PrInCE a unique resource for discovery.
For a detailed introduction to PrInCE, see the vignette:
```
browseVignettes("PrInCE")
```