---
title: autonomics
subtitle: generifying and intuifying cross-platform omics analysis
---
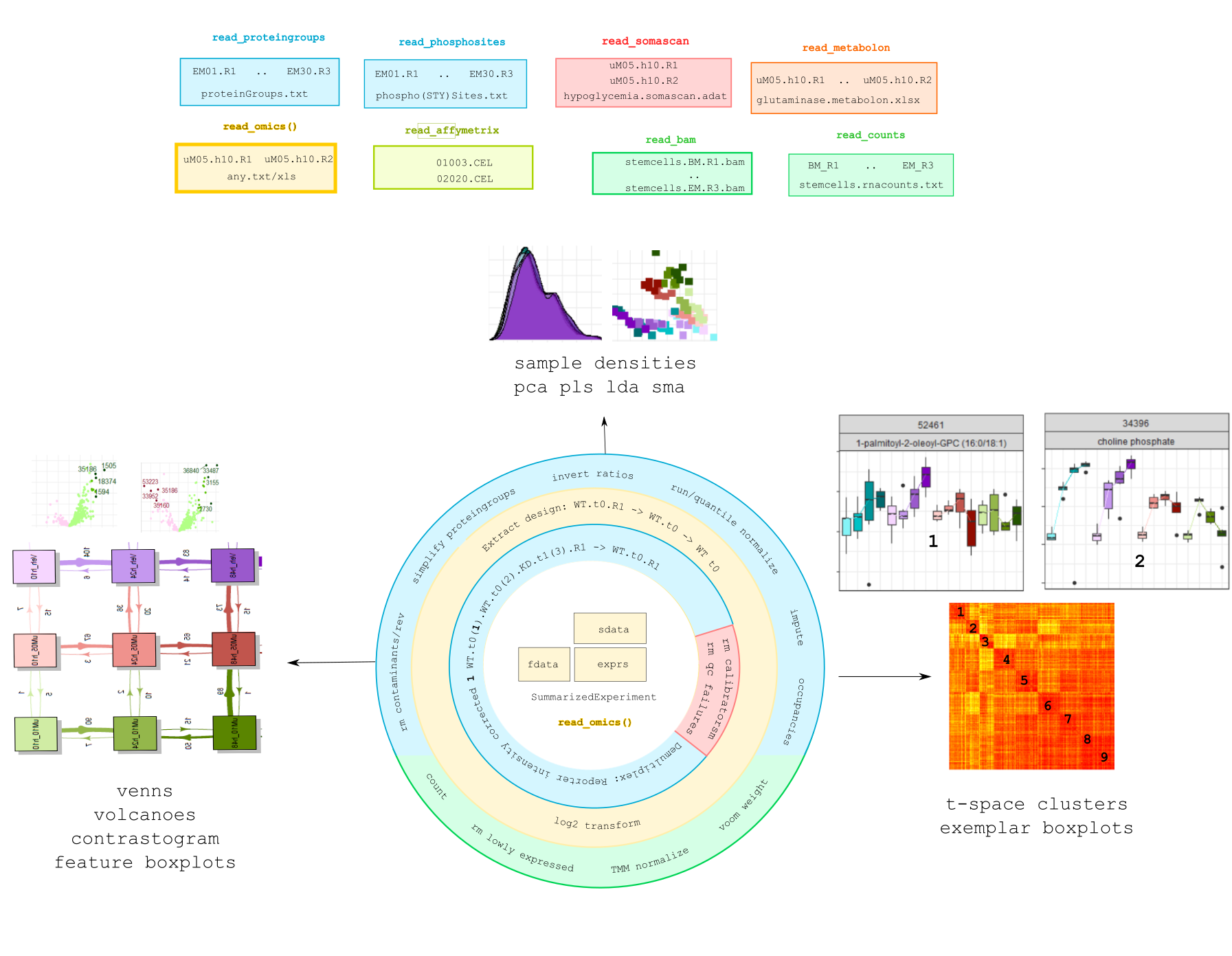
```
# Install
install.packages('remotes')
remotes::install_github('bhagwataditya/autonomics',
repos = BiocManager::repositories(),
dependencies = TRUE, upgrade = FALSE)
# Run
file <- download_data('billing16.bam.zip') # RNAseq BAM
read_rnaseq_bams(file, paired=TRUE, genome = 'hg38')
file <- download_data('billing16.rnacounts.txt') # RNAseq counts
read_rnaseq_counts(file)
file <- download_data('halama18.metabolon.xlsx') # Metabolon xlsx
read_metabolon(file)
select <- sprintf('%s_STD', c('E00', 'E01', 'E02', 'E05', 'E15', 'E30', 'M00'))
proteingroups <- download_data('billing19.proteingroups.txt') # LCMSMS Proteingroups
read_proteingroups(proteingroups, select_subgroups = select)
phosphosites <- download_data('billing19.phosphosites.txt') # LCMSMS Phosphosites
read_phosphosites(phosphosites, proteingroups, select = select_subgroups)
file <- download_data('atkin18.somascan.adat') # SOMAscan adat
read_somascan(file)
```