<!-- badges: start -->
[](https://www.nature.com/articles/s41467-024-44761-x)
[](https://www.tidyverse.org/lifecycle/#stable)
[](https://opensource.org/licenses/MIT)
[](https://github.com/js2264/HiContacts/actions/workflows/rworkflows.yml)
[](https://js2264.github.io/HiContacts)
[](https://js2264.github.io/OHCA/)
<a href=http://bioconductor.org/packages/release/bioc/html/HiContacts.html><img alt="Static Badge" src="https://img.shields.io/badge/Bioc_(release)-Landing_page-green?link=http%3A%2F%2Fbioconductor.org%2FcheckResults%2Fdevel%2Fbioc-LATEST%2FHiContacts%2F"></a>
<a href=http://bioconductor.org/checkResults/release/bioc-LATEST/HiContacts/><img alt="Bioc build (release)" src="https://img.shields.io/badge/dynamic/yaml?url=https%3A%2F%2Fbioconductor.org%2FcheckResults%2Frelease%2Fbioc-LATEST%2FHiContacts%2Fraw-results%2Fnebbiolo1%2Fbuildsrc-summary.dcf&query=%24.Status&label=Bioc%20build%20(release)&link=https%3A%2F%2Fbioconductor.org%2FcheckResults%2Frelease%2Fbioc-LATEST%2FHiContacts%2F"></a>
<a href=http://bioconductor.org/checkResults/devel/bioc-LATEST/HiContacts/><img alt="Bioc build (devel)" src="https://img.shields.io/badge/dynamic/yaml?url=https%3A%2F%2Fbioconductor.org%2FcheckResults%2Fdevel%2Fbioc-LATEST%2FHiContacts%2Fraw-results%2Fnebbiolo2%2Fbuildsrc-summary.dcf&query=%24.Status&label=Bioc%20build%20(devel)&link=https%3A%2F%2Fbioconductor.org%2FcheckResults%2Fdevel%2Fbioc-LATEST%2FHiContacts%2F"></a>
<!-- badges: end -->
# HiContacts
[👉 OHCA book 📖](https://js2264.github.io/OHCA/)
*Please cite:*
Serizay J, Matthey-Doret C, Bignaud A, Baudry L, Koszul R (2024). “Orchestrating chromosome conformation capture analysis with Bioconductor.” _Nature Communications_, **15**, 1-9. [doi:10.1038/s41467-024-44761-x](https://doi.org/10.1038/s41467-024-44761-x).
[](https://www.nature.com/articles/s41467-024-44761-x)
---
HiContacts provides tools to investigate `(m)cool` matrices imported in R by `HiCExperiment`.
It leverages the `HiCExperiment` class of objects, built on pre-existing Bioconductor objects, namely `InteractionSet`, `GInterations` and `ContactMatrix` (`Lun, Perry & Ing-Simmons, F1000Research 2016`), and provides **analytical** and **visualization** tools to investigate contact maps.
## Installation
`HiContacts` is available in Bioconductor. To install the current release, use:
```r
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("HiContacts")
```
To install the most recent version of `HiContacts`, you can use:
```r
install.packages("devtools")
devtools::install_github("js2264/HiContacts")
library(HiContacts)
```
## Citation
If you are using `HiContacts` in your research, please cite:
> Serizay J (2022). _HiContacts: HiContacts: R interface to cool files_.
> R package version 1.1.0
> <https://github.com/js2264/HiContacts>.
## How to use `HiContacts`
`HiContacts` includes a introduction vignette where its usage is
illustrated. To access the vignette, please use:
```r
vignette('HiContacts')
```
## Visualising Hi-C contact maps and features
### Importing a Hi-C contact maps file with `HiCExperiment`
```r
mcool_file <- HiContactsData::HiContactsData('yeast_wt', format = 'mcool')
range <- 'I:20000-80000' # range of interest
availableResolutions(mcool_file)
hic <- HiCExperiment::import(mcool_file, format = 'mcool', focus = range, resolution = 1000)
hic
```
### Plotting matrices (square or horizontal)
```r
plotMatrix(hic, use.scores = 'count')
plotMatrix(hic, use.scores = 'balanced', limits = c(-4, -1))
plotMatrix(hic, use.scores = 'balanced', limits = c(-4, -1), maxDistance = 100000)
```
### Plotting matrices with topological features
```r
library(rtracklayer)
mcool_file <- HiContactsData::HiContactsData('yeast_wt', format = 'mcool')
hic <- import(mcool_file, format = 'mcool', focus = 'IV')
loops <- system.file("extdata", 'S288C-loops.bedpe', package = 'HiContacts') |>
import() |>
InteractionSet::makeGInteractionsFromGRangesPairs()
borders <- system.file("extdata", 'S288C-borders.bed', package = 'HiContacts') |>
import()
p <- plotMatrix(
hic, loops = loops, borders = borders,
limits = c(-4, -1), dpi = 120
)
```
### Plotting aggregated matrices (a.k.a. APA plots)
```r
contacts <- contacts_yeast()
contacts <- zoom(contacts, resolution = 2000)
aggr_centros <- aggregate(contacts, targets = topologicalFeatures(contacts, 'centromeres'))
plotMatrix(aggr_centros, use.scores = 'detrended', limits = c(-1, 1), scale = 'linear')
```
## Mapping topological features
### Chromosome compartments
```r
microC_mcool <- fourDNData::fourDNData('4DNES14CNC1I', 'mcool')
hic <- import(microC_mcool, format = 'mcool', resolution = 10000000)
genome <- BSgenome.Mmusculus.UCSC.mm10::BSgenome.Mmusculus.UCSC.mm10
# - Get compartments
hic <- getCompartments(
hic, resolution = 100000, genome = genome, chromosomes = c('chr17', 'chr19')
)
# - Export compartments as bigwig and bed files
export(IRanges::coverage(metadata(hic)$eigens, weight = 'eigen'), 'microC_compartments.bw')
export(
topologicalFeatures(hic, 'compartments')[topologicalFeatures(hic, 'compartments')$compartment == 'A'],
'microC_A-compartments.bed'
)
export(
topologicalFeatures(hic, 'compartments')[topologicalFeatures(hic, 'compartments')$compartment == 'B'],
'microC_B-compartments.bed'
)
# - Generate saddle plot
plotSaddle(hic)
```
### Diamond insulation score and chromatin domains borders
```r
# - Compute insulation score
hic <- refocus(hic, 'chr19:1-30000000') |>
zoom(resolution = 10000) |>
getDiamondInsulation(window_size = 100000) |>
getBorders()
# - Export insulation as bigwig track and borders as bed file
export(IRanges::coverage(metadata(hic)$insulation, weight = 'insulation'), 'microC_insulation.bw')
export(topologicalFeatures(hic, 'borders'), 'microC_borders.bed')
```
## In-depth analysis of `HiCExperiment` objects
### Arithmetics
#### Detrend
#### Autocorrelate
#### Divide
#### Merge
### Distance law, a.k.a. P(s)
```r
hic <- import(CoolFile(
mcool_file,
pairs = HiContactsData::HiContactsData('yeast_wt', format = 'pairs.gz')
))
ps <- distanceLaw(hic)
plotPs(ps, ggplot2::aes(x = binned_distance, y = norm_p))
```
### Virtual 4C
```r
hic <- import(CoolFile(mcool_file))
v4C <- virtual4C(hic, viewpoint = GRanges('V:150000-170000'))
plot4C(v4C)
```
### Cis-trans ratios
```r
hic <- import(CoolFile(mcool_file))
cisTransRatio(hic)
```
### Scalograms
```r
```
## HiCExperiment ecosystem
`HiCool` is integrated within the `HiCExperiment` ecosystem in Bioconductor.
Read more about the `HiCExperiment` class and handling Hi-C data in R
[here](https://github.com/js2264/HiCExperiment).
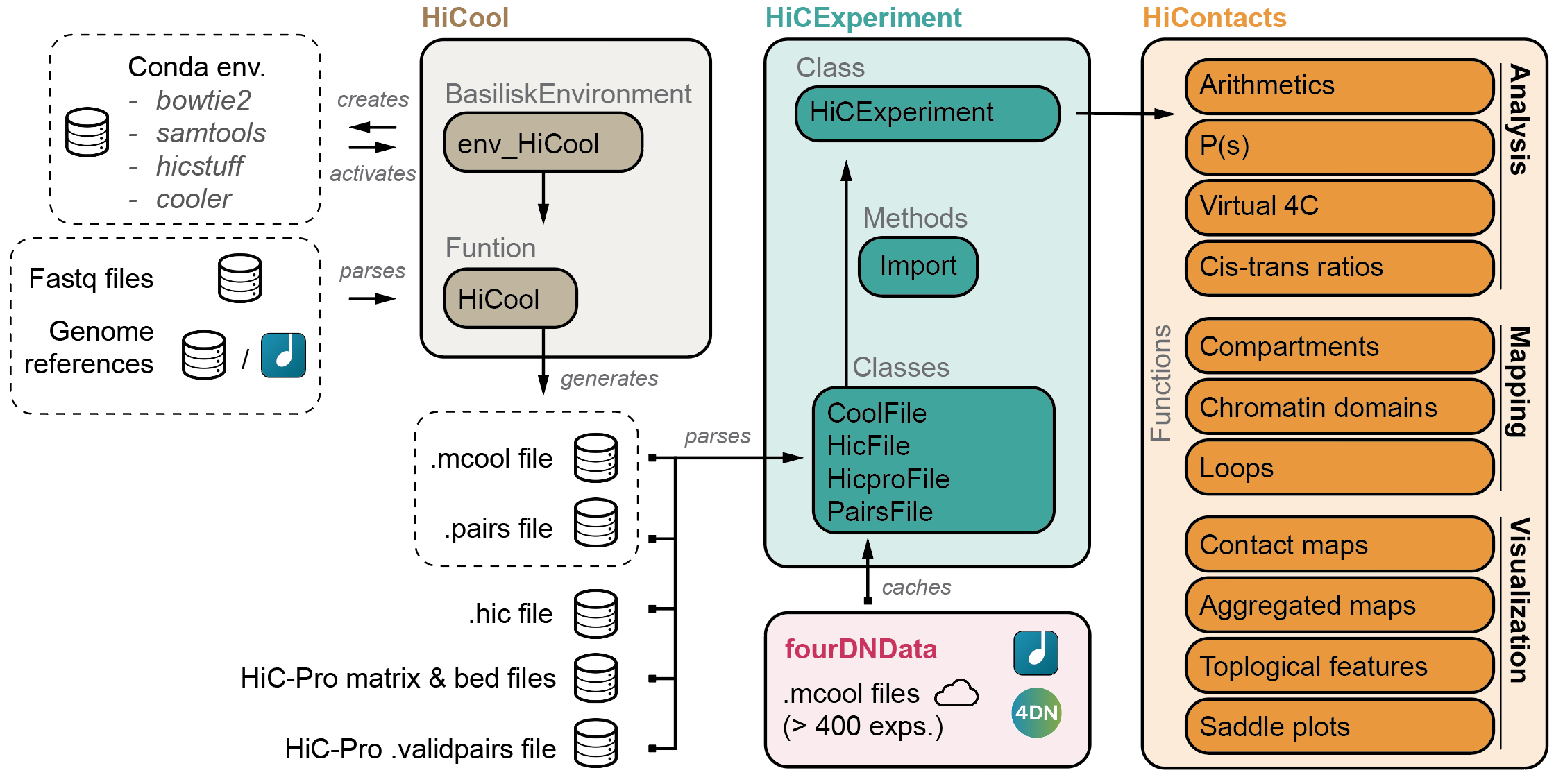
- [HiCExperiment](https://github.com/js2264/HiCExperiment): Parsing Hi-C files in R
- [HiCool](https://github.com/js2264/HiCool): End-to-end integrated workflow to process fastq files into .cool and .pairs files
- [HiContacts](https://github.com/js2264/HiContacts): Investigating Hi-C results in R
- [HiContactsData](https://github.com/js2264/HiContactsData): Data companion package
- [fourDNData](https://github.com/js2264/fourDNData): Gateway package to 4DN-hosted Hi-C experiments