# BrowserViz
BrowserViz provides the basis for, and a very simple working example
of, interactive R/browser visualization. Thus two interactive powerful
and complementary environments are linked, creating a powerful hybrid
setting for exploratory data analysis.
The basic architecture:
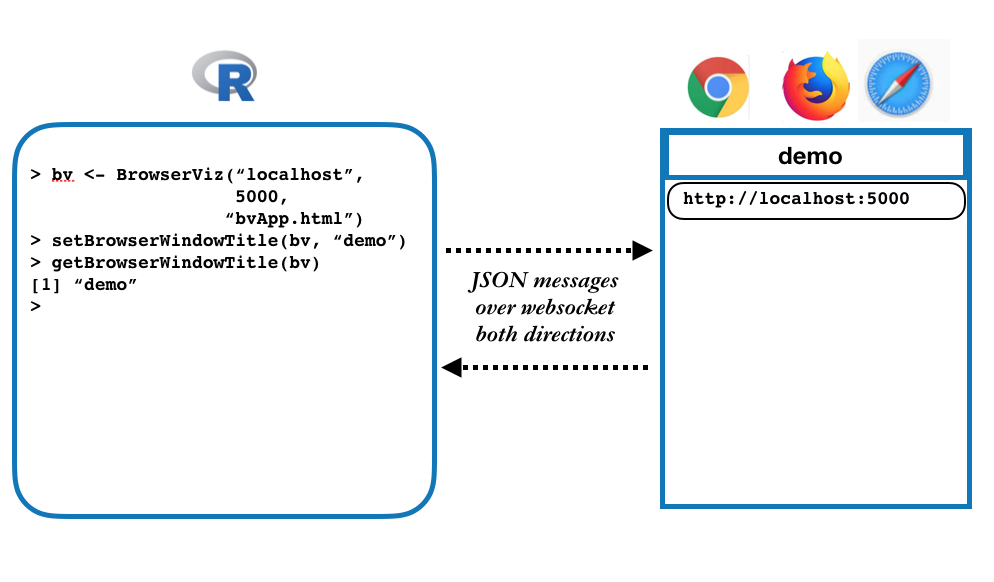
This work is motivated by our belief that contemporary web browsers,
supporting HTML5 and Canvas, and running increasingly powerful
Javascript libraries (for example, d3 and cytoscape.js) have become
the best setting in which to develop interactive graphics for
exploratory data analysis. We predict that web browsers, already
powerful and easily programmed, will steadily improve in rendiring
power and interactivity, and thus remain the optimal setting for
interactive R visualization for years to come.
BrowserViz is designed as an S4 **base** class, to be subclassed by
other packages. Two examples:
- igvR: a Bioconductor package with a high-level interface to the
igv.js genome browser
- RCyjs: a Bioconductor package for cytoscape.js
Both of these packages are intended for the interactive R user
(programmer, script writer, or novice bioinformatician) doing
exploratory data analysis, in need of visualzation of their data.
We also offer Shiny widget versions of these two Javascript libraries
for those creating point-and-click applications for the
non-programming user:
- igvShiny
- cyjShiny
We hope that others will write BrowserViz subclasses bringing other
Javascript visualization capability to the R user.
Though BrowserViz is a base class, it is concrete, not abstract - by
which I mean that you can create a BrowserViz object that is fully
functional, if limited in its capabilities. One of these
capabilities is used daily in testing the package. After creating a
BrowerViz ("bv") instance in R with a single line of code, these
methods can be called
- setBrowserWindowTitle(bv, "new title")
- getBrowserWindowTitle(bv)
- getBrowserInfo(bv)
- roundTripTest(bv, someRDataStructure) # e.g., LETTERS, mtcars